Nature-inspired compounds chop up cancer gene’s RNA
The cancer gene MYC drives unrestrained growth of most human cancers. It has been called the “Mount Everest” of cancer research because of the difficulty of designing medications that can disable it, and the expectation that an effective MYC drug could help so many cancer patients. Research groups from the The Wertheim UF Scripps Institute in Florida, the Max Planck Institute of Molecular Physiology in Dortmund and the University of Münster have joined forces and climbed that peak by developing Nature-inspired compounds chopping up MYC’s RNA. This innovative RNA degrader approach could also open new routes to summit other similarly hard-to-treat diseases.
How cancer cells grow, divide and spread is driven to a large extent by certain proteins, which makes them an important target in cancer research and therapy. However, most of the cancer-driving proteins have eluded all attempts by researchers to stop their harmful effects. Targeting cancer genes’ RNA offers an alternate route to tackle these diseases whose key protein structures couldn’t be reached by medications. RNA has many functions: beyond that of a protein-coding template, it regulates homeostasis in healthy and cancer cells through critical interactions with a plethora of cellular molecules. However, RNAs’ structures are so diverse and changeable that many in the pharmaceutical industry wrote off trying to make RNA-directed medicines as a pointless exercise.
Targeting RNA to Stop Cancer
Over the course of 15 years, Matthew Disney and his group have managed to identify many conserved, druggable RNA structures. Disney’s team built their own compound collections and showed in mice that targeting RNAs can wipe out cancer tumors and improve other diseases, including ALS and myotonic dystrophy. In 2016 during a scientific conference in Spain, when Waldmann and Disney compared notes after a talk, the idea for the partnership first arose. Both agreed that targeting RNA with drugs presented an exciting possible way to target incurable diseases, but it was early days. Nothing like that existed in the market.
In 2018 two members of Disney’s lab visited Waldmann’s lab in Dortmund to use their screening methods and compound libraries including lots of nature-inspired compounds to hunt for potential RNA-binding candidates. Disney said the results surpassed his expectations: they found the universe of known, druggable RNA structures had expanded dramatically. “We discovered around 2,000 new RNA structures that are able to bind drug-like small molecules, and identified six new chemotypes able to bind RNA,” Disney said. “We basically have created an encyclopedia of druggable RNA folds.”
Some of the compounds in the library were designed by the team of organic chemist Frank Glorius. “We develop molecules that exhibit sought-after-function in diverse biological areas, like the compounds in the library, that were originally designed to specifically influence biology in cell membranes. But we would have never expected them to bind RNA-structures.” Glorius said. The compounds derived from imidazole, a molecule common in natural products and drugs, modified with carbon-based chains, ultimately proved to be the most effective at binding to the cancer-linked RNAs MYC, JUN and microRNA-155.
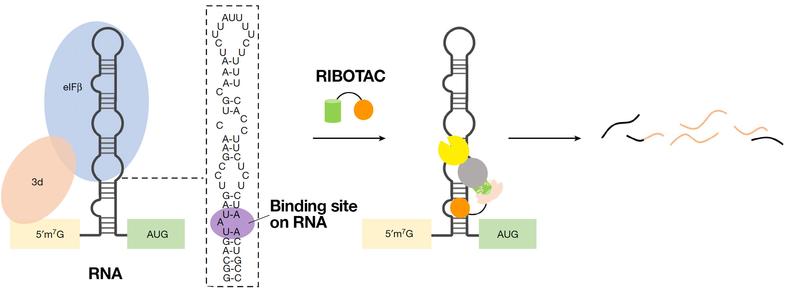
RNA Killer
However, binding these targets’ RNA alone didn’t make enough of an impact. So Disney’s team devised an innovative method for editing out these disease-causing RNA segments. He did so by attaching a chemical fishing hook to the molecules, one designed to catch the cell’s RNA recycling enzymes. It worked as planned. The RNA recycling enzyme chopped up the RNA that the drug molecule was attached to, preventing the disease-causing proteins from being built. They called their hybrid molecule a RiboTAC, short for “ribonuclease targeting chimera.”
“With the degrader added, we started seeing these ‘undruggable’ cancer RNAs reduced by 35%, 40%, 50% or more. This caused cancer cells to die and cleared tumors in mouse-based studies of breast cancer that spread to lungs,” Disney said.
“This study shows that compound classes inspired by natural products provide a rich, new source for targeting RNA disease targets in general. Of course, we are developing them further, following our concept of pseudo natural products” Waldmann says. Because the strategy is so novel, it will take several years of additional work before the innovations reach patients via a clinical trial, he added. “This is a long road to take, a marathon run, actually,” Waldmann says.
“The compounds are a good starting point and show us where to go to build small molecules, RNA-targeting medicines that could eventually treat patients with diseases like aggressive cancers that currently have poor or no options,” Disney say. “This new data also shows us that this approach could have many other disease applications.”
Originalpublikation:
Tong Y, Lee Y, Liu X, Childs-Disney JL, Suresh BM, Benhamou RI, Yang C, Li W, Costales MG, Haniff HS, Sievers S, Abegg D, Wegner T, Paulisch TO, Lekah E, Grefe M, Crynen G, Van Meter M, Wang T, Gibaut QMR, Cleveland JL, Adibekian A, Glorius F, Waldmann H, Disney MD (2023). Programming inactive RNA-binding small molecules into bioactive degraders.
Nature https://doi.org/10.1038/s41586-023-06091-8
Weitere Informationen:
https://www.mpi-dortmund.mpg.de/news/rna-killer-stop-cancer
Ähnliche Pressemitteilungen im idw