A map to study the behavior of early pancreas cells
Dresden researchers connect imaging and genetic data to gain insights into the development of the pancreas.
Our pancreas has different cells that do important jobs to control our blood sugar. One of the genes in pancreatic cells is called Neurogenin 3 (NEUROG3). Its mutation can lead to Diabetes. It is active only for a short period of time during the development of the pancreas, which is why its behavior and dynamics have remained a mystery, especially in the context of human development. Researchers from the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany, and Novo Nordisk Foundation at University of Copenhagen, Denmark, used a special method to watch both the gene's activity and the protein it makes in human pancreas cells to better understand the gene. The researchers developed a methodology that can link the dynamic behaviors of pancreatic cells observed in live imaging movies to all the genes they express. This will contribute to a better understanding of how the hormone-producing cells of the pancreas develop and could pave the way to produce more of these cells for therapeutic purposes, such as the production and transplantation of these cells to patients suffering from diabetes.
The different cells in the pancreas control our blood sugar, such as beta cells that make insulin. Insulin helps lower our blood sugar. If these cells stop working or die, we can get sick with diabetes. When our body is growing, all these special cells come from a single cell type in the pancreas, which is called a pancreatic endocrine progenitor. This cell uses a gene called NEUROG3 for a short time to do its job.
The research group of Anne Grapin-Botton, managing director at the MPI-CBG in Dresden, together with colleagues from the Novo Nordisk Foundation at University of Copenhagen, set out to learn more about these special cells in the pancreas that use the gene NEUROG3 and to see how this gene behaves in single cells.
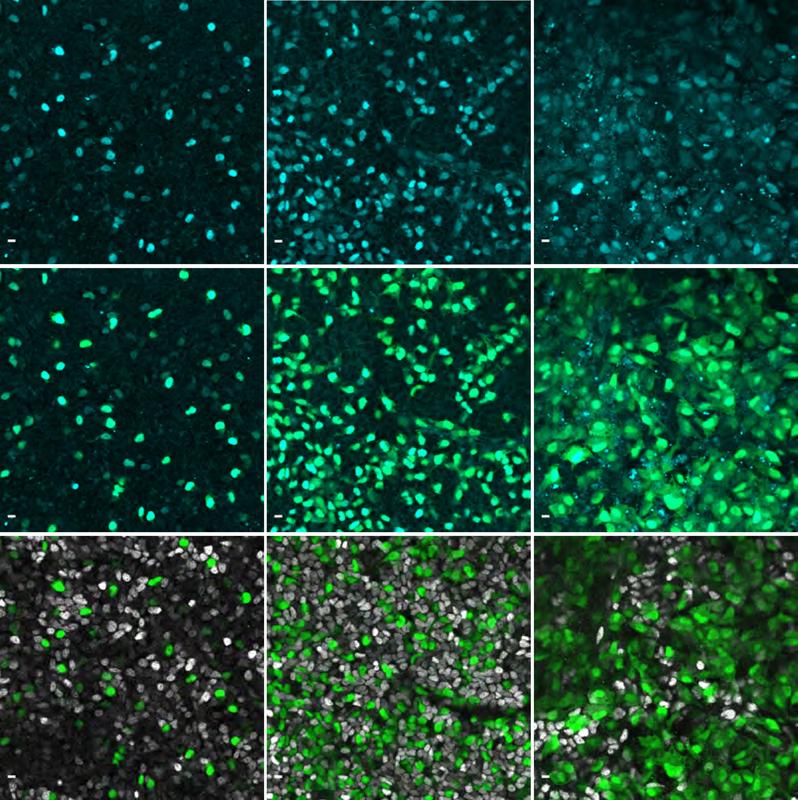
“We used special tags to see NEUROG3 in these cells and watched how they move using a long-term live imaging method that generated videos,” explains Belin Selcen Beydag-Tasöz, the first author of the study, and continues: “By looking at flat 2D and 3D models of human pancreas growth, we found out that the levels of the NEUROG3 gene were different in different cells. Some cells had a lot of this gene, and some had a little. Surprisingly, despite this heterogeneity, all the cells that had detectable NEUROG3 formed cells producing hormones. Another surprising result was that NEUROG3 works about two times slower in humans than in mice, meaning it takes more time for the gene to do its job in humans compared to mice.”
The researchers used the long-term live imaging method to see a process normally hidden in the mother’s womb. The brightness of the cells helped them combine the gene activity with how the cells were behaving. Using this method, the research team learned that another gene called KLK12 has a role in making cells move to start forming islets of Langerhans when the NEUROG3 gene starts working.
Anne Grapin-Botton, who supervised the study, summarizes: “The cell culture systems we developed to understand how cells in human fetuses form organs are starting to bear fruit. In our study, we've learned a lot more about how certain genes activity during fetal development can lead to diabetes later in life. The results show that when producing endocrine cells for future therapeutic applications based on the transplantation of these cells into diabetic patients, there is some flexibility in the control of NEUROG3.”
Wissenschaftlicher Ansprechpartner:
Dr. Anne Grapin-Botton
+49 (0) 351 210-2500
botton@mpi-cbg.de
Originalpublikation:
Belin Selcen Beydag-Tasöz, Joyson Verner D’Costa, Lena Hersemann, Byung Ho Lee, Federica Luppino, Yung Hae Kim, Christoph Zechner, Anne Grapin-Botton: Integrating single-cell imaging and RNA sequencing datasets links differentiation and morphogenetic dynamics of human pancreatic endocrine progenitors, Developmental Cell, 2023, https://doi.org/10.1016/j.devcel.2023.07.019.
Ähnliche Pressemitteilungen im idw