Scientists discover how yew trees save lives
Researchers from the Max Planck Institute of Molecular Plant Physiology unravel the biosynthetic pathway of paclitaxel in Yew plants, a most successfully used chemotherapeutic for cancer treatment. This discovery might facilitate the production of this very complex molecule which is currently produced with great efforts and high costs.
Paclitaxel helps fight cancer and is made from yew
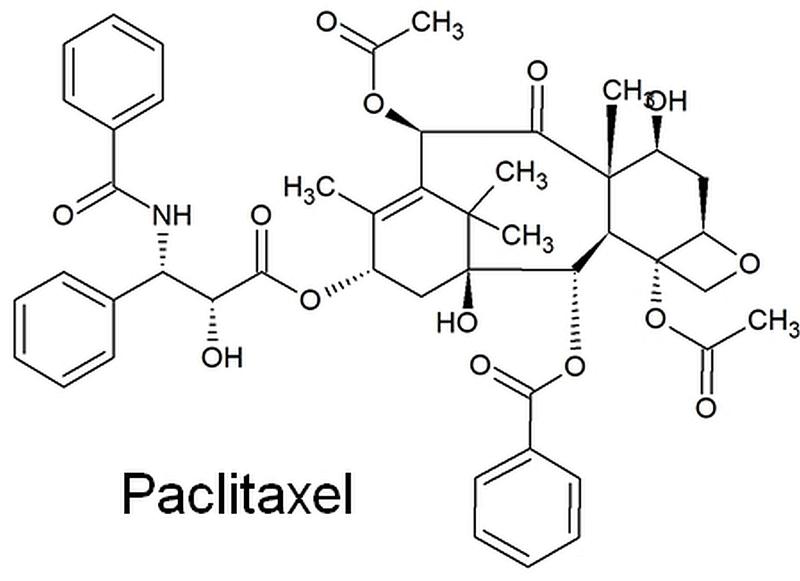
Cancer in all its forms is still one of the most common diseases and very hard to treat. Part of modern cancer therapy is the use of toxic chemicals, called chemotherapeutics, that kill the tumor. Unfortunately, these chemicals are often very complex, difficult to obtain and thus expensive. The very successful chemotherapeutic paclitaxel is branded as Taxol®. It is a natural plant remedy derived from yew trees. As 10.000 kg of yew needles need to be extracted to obtain a 2g dosage for treatment of a single patient, other methods of production are desperately sought for to make this drug more accessible to the people in need. Artificial synthesis of paclitaxel in the lab is possible, but due to its complicated chemical structure, it requires many complex steps and is thus even more expensive than extracting it from truckloads of needles and bark.
Enzymes allow plants to build complex molecules
The yew plant produces this complex chemical using enzymes. These enzymes are tools that facilitate this long chain of chemical reactions that lead to the final paclitaxel molecule. It is free of charge, but the plant produces only minute amounts of this compound. A commonly used way to speed up drug production in such cases would be to copy the genetic code for these enzymes and to move them to another organism, a bacterium for example or to a plant that can easily be grown in large quantities and from which the drug can then be extracted easily and in larger amounts than from yew needles. This approach is used for insulin production for example. However, to do this, scientists need to know all the enzymes and their genetic code to copy the process in question. For paclitaxel biosynthesis a lot of the enzymes and intermediate products were unknown.
The missing links in the chain are like needles in a haystack
Youjun Zhang and colleagues from the Max Planck Institute of Molecular Plant Physiology were now able to identify all the missing steps required to produce paclitaxel in plants at once. They analyzed data from twelve experiments that involved data from several ten thousand genes in yew plants to find sequences for enzymes that were produced in similar amounts as the few other enzymes already known to be involved in the paclitaxel pathway. Using sophisticated chemical analyses and molecular biological tools, they were able to reproduce the whole biosynthetic pathway from yew plants and to copy all the enzymes into the Australian tobacco relative Nicotiana benthamiana. These transgenic Nicotiana plants indeed produced similar amounts of paclitaxel as yew. The researchers also tried to copy the paclitaxel pathway into bacteria, but found that some of the enzymes just don`t work in bacterial cells.
“It might have to do with the location of the enzymes inside the plant cells. In plants, most of the enzymes involved in paclitaxel biosynthesis are fixed to a specific membrane. Thus, they are close together potentially building a transport chain in which each enzyme takes over the product from the one before and modifies it a bit further until at the end the final paclitaxel is released. As bacteria have different membranes than plants, the enzymes may just not find each other.” Says Youjun Zhang the lead author of the study.
The fact that now the whole biosynthetic pathway for one of the worlds most successful chemotherapeutics is unveiled bears the potential to find ways to speed up paclitaxel production. Alisdair Fernie, the head of the research group believes that “the discovery of the complete paclitaxel biosynthesis pathway opens the door for applied research on optimizing its production and it seems likely that it will be possible to adjust the process to work in bacteria as well, which would allow for large scale production. Until then, the Nicotiana bethamiana lines we created can be used to optimize the system and to increase the yield of existing production lines that use plant tissue cultures.”
Wissenschaftlicher Ansprechpartner:
Prof. Alisdair Fernie
Max Planck Institute of Molecular Plant Physiology
Potsdam Science Park
Am Mühlenberg 1
14476 Potsdam, Germany
Fernie@mpimp-golm.mpg.de
Originalpublikation:
Youjun Zhang, Lorenz Wiese, Hao Fang, Saleh Alseekh, Leonardo Perez de Souza, Federico Scossa, John Molloy, Mathias Christmann, Alisdair R Fernie, Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis, Molecular Plant, 4.12.2023, doi: https://doi.org/10.1016/j.molp.2023.10.016
Weitere Informationen:
https://doi.org/10.1016/j.molp.2023.10.016 - link to the original publication
https://www.mpimp-golm.mpg.de/2728818/4fernie - website of the research group
Ähnliche Pressemitteilungen im idw