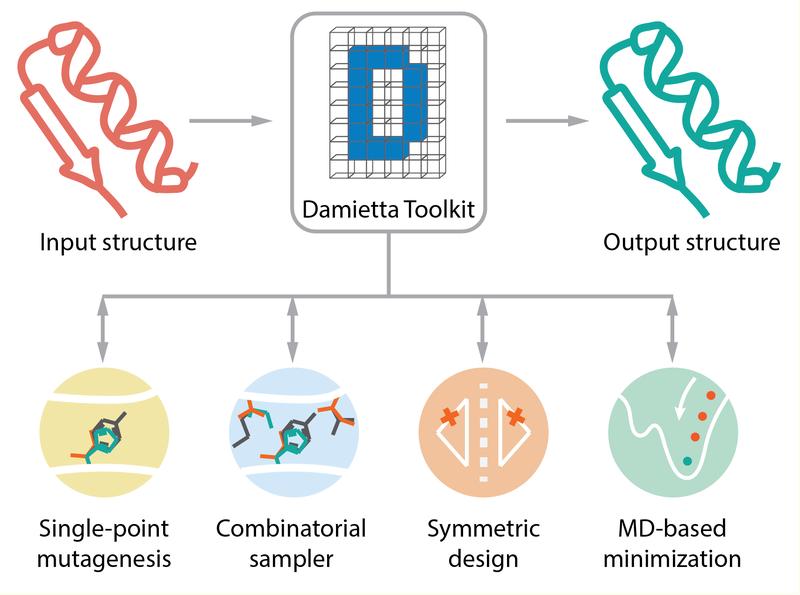
Toolkit makes protein design faster and more accessible
Researchers at the Max Planck Institute for Biology Tübingen, the University of Tübingen, and the University Hospital Tübingen have developed a web-based toolkit to accelerate and simplify protein design without needing powerful computers or extensive protein design expertise on the user’s end. The toolkit benefits its users with multiple design tools, fast analyses, easy interpretation, and downloadable results. Their framework for the Damietta Server, published in Nucleic Acids Research, offers a comprehensive resource for biological research community.
Designing proteins with novel functions is a crucial step in many areas of biomedical research. Protein design process has often been complex, requiring significant computing power and expertise, thus limiting its applicability and accessibility to a broader range of researchers.
The Damietta protein design toolkit (https://damietta.de) addresses these challenges by offering a web-based platform that integrates several protein design tools. Such a platform was inspired by the MPI bioinformatics toolkit (https://toolkit.tuebingen.mpg.de), which greatly simplifies bioinformatics workflows, and is hosted by the Department of Protein Evolution at the Max Planck Institute for Biology Tübingen, headed by Prof. Andrei Lupas, who supported the development of the new protein design toolkit.
"Damietta’s user-friendly interface makes it a powerful tool for researchers of all experience levels," says Dr. Kateryna Maksymenko, a researcher in the Department of Protein Evolution at the Max Planck Institute for Biology Tübingen. "This toolkit significantly accelerates protein design, boosting discoveries and direct clinical applications in biomedicine."
Unlike machine learning-based methods, Damietta relies on well-established physics principles, ensuring generalisable, accurate, and interpretable results.
The Damietta Server simplifies the process of protein design for users by offering:
• Faster and more accurate calculations. What used to take weeks can now be completed in a day or less.
• Intuitive graphical interface, allowing researchers with limited protein design experience to use the toolkit effectively.
• Improved transparency by providing clear and interpretable results, enabling researchers to understand the underlying physical interactions influencing the designed proteins.
Wider potential for new biomedical and therapeutic applications
Previously, Damietta software demonstrated its effectiveness by designing two classes of proteins for cancer treatment. The team also uses the software to develop proteins that are helpful for various disorders, including bone marrow failure and leukaemia.
Current users can look forward to future updates, as the researchers are actively developing Damietta to enable setting up membrane environment and to incorporate non-standard amino acids and small molecules.
The free web server, interactive tutorials and user manuals are available at https://damietta.de
Wissenschaftlicher Ansprechpartner:
Dr. Mohammad ElGamacy
mohammad.elgamacy@med.uni-tuebingen.de
Originalpublikation:
Grin I, Maksymenko K, Wörtwein T & ElGamacy M (2024): The Damietta Server: a comprehensive protein design toolkit. Nucleic Acids Res, gkae297, 25 April 2024, https://doi.org/10.1093/nar/gkae297
Weitere Informationen:
https://damietta.de
https://www.bio.mpg.de/330048/news_publication_22000296_transferred?c=57217
https://doi.org/10.1093/nar/gkae297
Die semantisch ähnlichsten Pressemitteilungen im idw